In Depth: Parameter tuning for Random Forest
In this post we will explore the most important parameters of Random Forest and how they impact our model in term of overfitting and underfitting.
A random forest is a meta estimator that fits a number of decision tree classifiers on various sub-samples of the dataset and use averaging to improve the predictive accuracy and control over-fitting.
We will use the Titanic Data from kaggle. For the sake of this post, we will perform as little feature engineering as possible as it is not the purpose of this post.
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
Load train data
# get titanic & test csv files as a DataFrame
train = pd.read_csv("input/train.csv")
print train.shape
(891, 12)
train.head()
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
Check for missing values
#Checking for missing data
NAs = pd.concat([train.isnull().sum()], axis=1, keys=['Train'])
NAs[NAs.sum(axis=1) > 0]
| Train | |
|---|---|
| Age | 177 |
| Cabin | 687 |
| Embarked | 2 |
We will remove ‘Cabin’, ‘Name’ and ‘Ticket’ columns as they require some processing to extract useful features
# At this point we will drop the Cabin feature since it is missing a lot of the data
train.pop('Cabin')
# At this point names don't affect our model so we drop it
train.pop('Name')
# At this point we drop Ticket feature
train.pop('Ticket')
train.shape
(891, 9)
Fill the missing age values by the mean value
# Filling missing Age values with mean
train['Age'] = train['Age'].fillna(train['Age'].mean())
Fill the missing ‘Embarked’ values by the most frequent value
# Filling missing Embarked values with most common value
train['Embarked'] = train['Embarked'].fillna(train['Embarked'].mode()[0])
train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Fare | Embarked | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | male | 22.0 | 1 | 0 | 7.2500 | S |
| 1 | 2 | 1 | 1 | female | 38.0 | 1 | 0 | 71.2833 | C |
| 2 | 3 | 1 | 3 | female | 26.0 | 0 | 0 | 7.9250 | S |
| 3 | 4 | 1 | 1 | female | 35.0 | 1 | 0 | 53.1000 | S |
| 4 | 5 | 0 | 3 | male | 35.0 | 0 | 0 | 8.0500 | S |
‘Pclass’ is a categorical feature so we convert its values to strings
train['Pclass'] = train['Pclass'].apply(str)
Let’s perform a basic one hot encoding of categorical features
# Getting Dummies from all other categorical vars
for col in train.dtypes[train.dtypes == 'object'].index:
for_dummy = train.pop(col)
train = pd.concat([train, pd.get_dummies(for_dummy, prefix=col)], axis=1)
train.head()
| PassengerId | Survived | Age | SibSp | Parch | Fare | Pclass_1 | Pclass_2 | Pclass_3 | Sex_female | Sex_male | Embarked_C | Embarked_Q | Embarked_S | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 22.0 | 1 | 0 | 7.2500 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 |
| 1 | 2 | 1 | 38.0 | 1 | 0 | 71.2833 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| 2 | 3 | 1 | 26.0 | 0 | 0 | 7.9250 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 |
| 3 | 4 | 1 | 35.0 | 1 | 0 | 53.1000 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| 4 | 5 | 0 | 35.0 | 0 | 0 | 8.0500 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 |
labels = train.pop('Survived')
For testing, we choose to split our data to 75% train and 25% for test
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(train, labels, test_size=0.25)
Let’s first fit a random forest with default parameters to get a baseline idea of the performance
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier()
rf.fit(x_train, y_train)
RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_split=1e-07, min_samples_leaf=1,
min_samples_split=2, min_weight_fraction_leaf=0.0,
n_estimators=10, n_jobs=1, oob_score=False, random_state=None,
verbose=0, warm_start=False)
y_pred = rf.predict(x_test)
We will use AUC (Area Under Curve) as the evaluation metric. Our target value is binary so it’s a binary classification problem. AUC is a good way for evaluation for this type of problems.
from sklearn.metrics import roc_curve, auc
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
roc_auc
0.76481643356643347
N_estimators
n_estimators represents the number of trees in the forest. Usually the higher the number of trees the better to learn the data. However, adding a lot of trees can slow down the training process considerably, therefore we do a parameter search to find the sweet spot.
n_estimators = [1, 2, 4, 8, 16, 32, 64, 100, 200]
train_results = []
test_results = []
for estimator in n_estimators:
rf = RandomForestClassifier(n_estimators=estimator, n_jobs=-1)
rf.fit(x_train, y_train)
train_pred = rf.predict(x_train)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_train, train_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
train_results.append(roc_auc)
y_pred = rf.predict(x_test)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
test_results.append(roc_auc)
from matplotlib.legend_handler import HandlerLine2D
line1, = plt.plot(n_estimators, train_results, 'b', label="Train AUC")
line2, = plt.plot(n_estimators, test_results, 'r', label="Test AUC")
plt.legend(handler_map={line1: HandlerLine2D(numpoints=2)})
plt.ylabel('AUC score')
plt.xlabel('n_estimators')
plt.show()
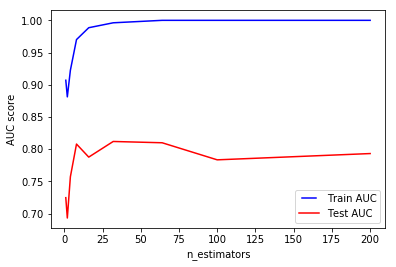
We can see that for our data, we can stop at 32 trees as increasing the number of trees decreases the test performance.
max_depth
max_depth represents the depth of each tree in the forest. The deeper the tree, the more splits it has and it captures more information about the data. We fit each decision tree with depths ranging from 1 to 32 and plot the training and test errors.
max_depths = np.linspace(1, 32, 32, endpoint=True)
train_results = []
test_results = []
for max_depth in max_depths:
rf = RandomForestClassifier(max_depth=max_depth, n_jobs=-1)
rf.fit(x_train, y_train)
train_pred = rf.predict(x_train)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_train, train_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
train_results.append(roc_auc)
y_pred = rf.predict(x_test)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
test_results.append(roc_auc)
from matplotlib.legend_handler import HandlerLine2D
line1, = plt.plot(max_depths, train_results, 'b', label="Train AUC")
line2, = plt.plot(max_depths, test_results, 'r', label="Test AUC")
plt.legend(handler_map={line1: HandlerLine2D(numpoints=2)})
plt.ylabel('AUC score')
plt.xlabel('Tree depth')
plt.show()
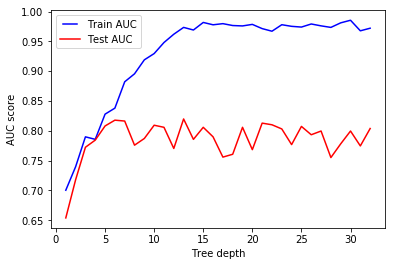
We see that our model overfits for large depth values. The trees perfectly predicts all of the train data, however, it fails to generalize the findings for new data
min_samples_split
min_samples_split represents the minimum number of samples required to split an internal node. This can vary between considering at least one sample at each node to considering all of the samples at each node. When we increase this parameter, each tree in the forest becomes more constrained as it has to consider more samples at each node. Here we will vary the parameter from 10% to 100% of the samples
min_samples_splits = np.linspace(0.1, 1.0, 10, endpoint=True)
train_results = []
test_results = []
for min_samples_split in min_samples_splits:
rf = RandomForestClassifier(min_samples_split=min_samples_split)
rf.fit(x_train, y_train)
train_pred = rf.predict(x_train)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_train, train_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
train_results.append(roc_auc)
y_pred = rf.predict(x_test)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
test_results.append(roc_auc)
from matplotlib.legend_handler import HandlerLine2D
line1, = plt.plot(min_samples_splits, train_results, 'b', label="Train AUC")
line2, = plt.plot(min_samples_splits, test_results, 'r', label="Test AUC")
plt.legend(handler_map={line1: HandlerLine2D(numpoints=2)})
plt.ylabel('AUC score')
plt.xlabel('min samples split')
plt.show()
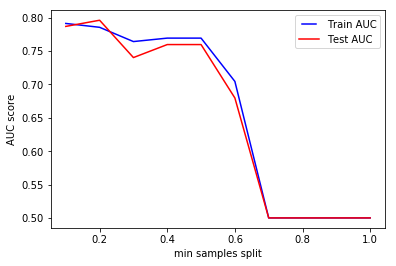
We can clearly see that when we require all of the samples at each node, the model cannot learn enough about the data. This is an underfitting case.
min_samples_leaf
min_samples_leaf is The minimum number of samples required to be at a leaf node. This parameter is similar to min_samples_splits, however, this describe the minimum number of samples of samples at the leafs, the base of the tree.
min_samples_leafs = np.linspace(0.1, 0.5, 5, endpoint=True)
train_results = []
test_results = []
for min_samples_leaf in min_samples_leafs:
rf = RandomForestClassifier(min_samples_leaf=min_samples_leaf)
rf.fit(x_train, y_train)
train_pred = rf.predict(x_train)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_train, train_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
train_results.append(roc_auc)
y_pred = rf.predict(x_test)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
test_results.append(roc_auc)
from matplotlib.legend_handler import HandlerLine2D
line1, = plt.plot(min_samples_leafs, train_results, 'b', label="Train AUC")
line2, = plt.plot(min_samples_leafs, test_results, 'r', label="Test AUC")
plt.legend(handler_map={line1: HandlerLine2D(numpoints=2)})
plt.ylabel('AUC score')
plt.xlabel('min samples leaf')
plt.show()
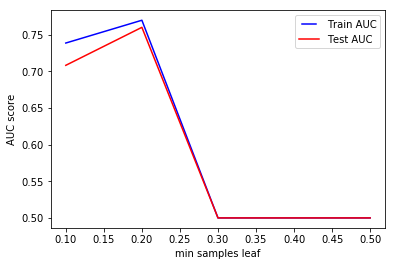
Same conclusion as to previous parameter. Increasing this value can cause underfitting.
max_features
max_features represents the number of features to consider when looking for the best split.
max_features = list(range(1,train.shape[1]))
train_results = []
test_results = []
for max_feature in max_features:
rf = RandomForestClassifier(max_features=max_feature)
rf.fit(x_train, y_train)
train_pred = rf.predict(x_train)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_train, train_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
train_results.append(roc_auc)
y_pred = rf.predict(x_test)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
test_results.append(roc_auc)
from matplotlib.legend_handler import HandlerLine2D
line1, = plt.plot(max_features, train_results, 'b', label="Train AUC")
line2, = plt.plot(max_features, test_results, 'r', label="Test AUC")
plt.legend(handler_map={line1: HandlerLine2D(numpoints=2)})
plt.ylabel('AUC score')
plt.xlabel('max features')
plt.show()
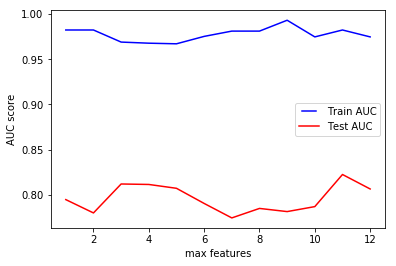
According to sklearn documentation for decision tree, the search for a split does not stop until at least one valid partition of the node samples is found, even if it requires to effectively inspect more than max_features features.
You can also check other tree based models like Decision Tree and Gradient Boosting